The KEGG identifies each reaction based on all participant compounds.
A SER REESCRITO PELO RODRIGO
However, some reactions are very similar, having the same principal substrates and
products. They differ only in secondary compounds.
, differing just in the
secondary compounds. These compounds are highlighted in red in Table 1. This can be a problem
when detecting the APs nodes because the pathway graph will have several pseudo alternatives
routes to bypass a specific point that could be a critical bottleneck.
For instance, we can take one reaction mediated by the enzyme ec:2.7.1.1, as seen in Figure
below, where four reactions provide the identical product from the same substrate
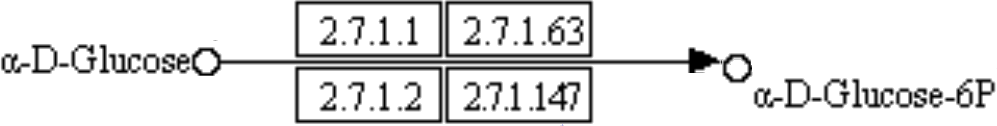
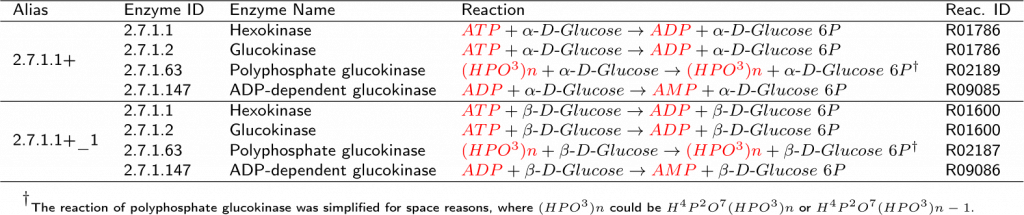
The KEGG canonical pathway is a compilation of many organisms, and some enzymes shown are exclusive for a taxon. In the example above, the enzyme 2.7.1.63 is not present in humans, being used by mycobacterial metabolism [1]. In other cases, the expression of a specific enzyme is tissue-dependent, being predominant in some organs. As an example, hexokinase (2.7.1.1 ) is active in mammalian glucose-dependent tissues, like the heart and brain, and glucokinase (2.7.1.2 ) is found predominantly in the liver [2, 3, 4]. The same occurs with the enzymes 5.1.3.9 and 5.3.1.15, where this last one is founded just in the yeast metabolism [5].
Thus, we need to convert the KEGG’s maps into a graph using a method capable of collapsing the redundant routes inside the pathways, reducing the inaccuracy in the APs detection.
[1] M. SZYMONA and W. OSTROWSKI, “INORGANIC POLYPHOSPHATE GLUCOKINASE OF MYCOBACTERIUM PHLEI,” Biochim Biophys Acta, vol. 85, pp. 283–295, May 1964.
[2] J. E. Wilson, Hexokinases, pp. 65–198. Berlin, Heidelberg: Springer Berlin Heidelberg, 1995.
[3] C. Postic, M. Shiota, and M. A. Magnuson, “Cell-specific roles of glucokinase in glucose homeostasis,” Recent Prog Horm Res, vol. 56, pp. 195–217, 2001.
[4] J. E. Wilson, “Isozymes of mammalian hexokinase: structure, subcellular localization and metabolic function,” J Exp Biol, vol. 206, pp. 2049–2057, Jun 2003.
[5] B. Wurster and B. Hess, “Glucose-6-phosphate-1-epimerase from baker’s yeast. A new enzyme,” FEBS Lett, vol. 23, pp. 341–344, Jul 1972.

