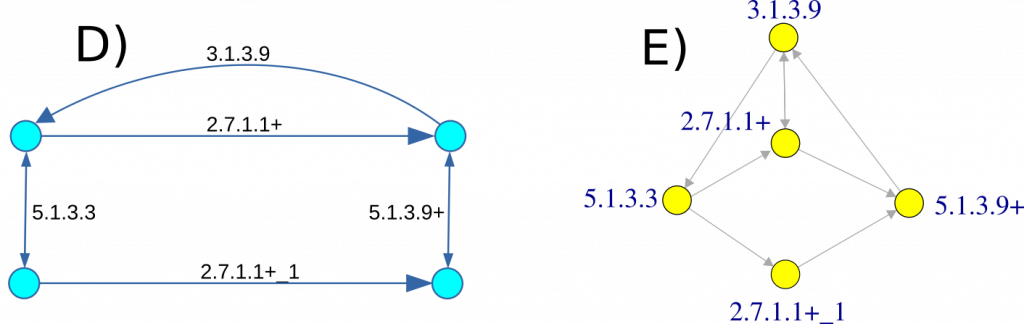
To enhance the accuracy and efficiency of metabolic network representation, we introduce a new entity, called gReaction, to serve as the edges in graph G. The classification of all reactions and enzymes reported in KEGG follows the criteria outlined below:
- If a set of reactions or enzymes utilize the same primary substrates and products and share the same reversibility properties, they are collectively classified as a single gReaction.
- If only one reaction or enzyme is present within a gReaction definition, it retains the exact name of the enzyme. For example, in our dataset, the enzymes 3.1.3.9 and 5.1.3.3 each correspond to a unique gReaction.
- If multiple reactions or enzymes fall under the same gReaction definition, the entity is named after the first enzyme in the group, followed by a “+” sign. For instance, the enzyme set {2.7.1.1, 2.7.1.2, 2.7.1.63, 2.7.1.147} is designated as 2.7.1.1+.
- If a single enzyme participates in multiple reactions, the gReaction name consists of the enzyme name appended with “_N,” where N represents a sequential identifier. For example, the enzyme ec:1.3.1.3 mediates eight distinct reactions (rn:R04823, rn:R04817, rn:R03713, rn:R02219, rn:R02841, rn:R02893, rn:R01835, rn:R02498). Consequently, the corresponding gReactions are assigned sequential aliases: {ec:1.3.1.3, ec:1.3.1.3_1, …, ec:1.3.1.3_6, ec:1.3.1.3_7}.